
Morgan Lab Research Initiatives
Morgan Lab researchers lead a number of research initiatives.
The Molecular Basis of Progression Multiple Myeloma
We have shown that myeloma is not a single disease but is rather composed of at least six distinct subsets with different biological features, each of which could be targeted therapeutically.
We have studied whole exome sequence data from monoclonal gammopathy of undetermined significance (MGUS), smoldering multiple myeloma (SMM), and newly-diagnosed multiple myeloma (NDMM) together with light chain amyloidosis and have defined the landscape of mutations, chromosomal translocations, structural events, and copy number abnormalities.
We have identified a distinct set of drivers within each of the disease subsets and identified the central importance of mutational activation of the RAS and NFkB pathways (see Figure 1).

Our studies have highlighted mutations that can be used clinically to define the prognostic outcome for individual patients or that can be targeted therapeutically.
We have described the landscape of complex rearrangements in the noncoding regions of the multiple myeloma genome including the complex events that include chromothripsis, chromoplexy, and templated insertions.
We are studying the impact of complex structural variants on the epigenetic control of gene regulation and how it could be targeted in multiple myeloma (see Figure 2).
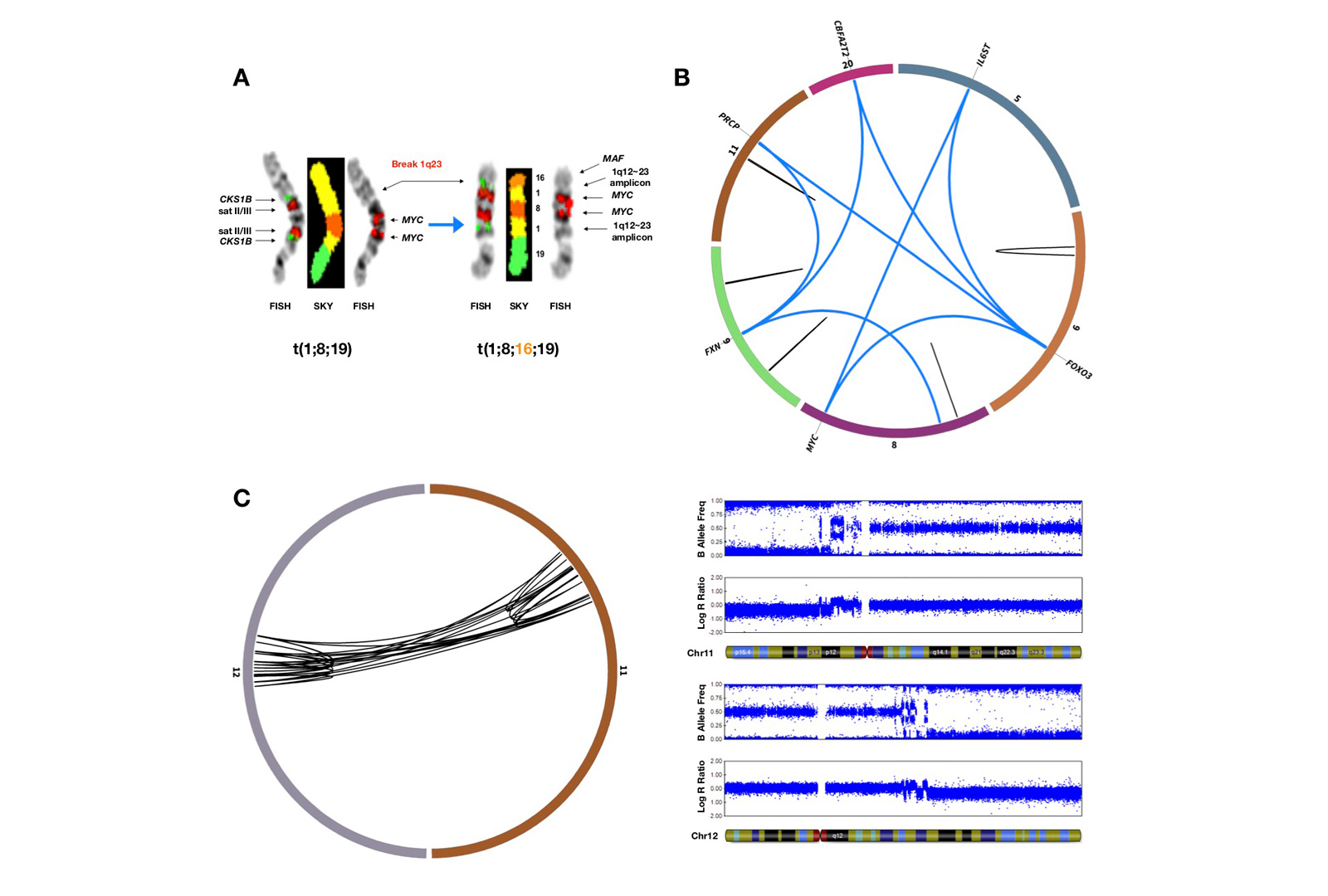
Other important genomic changes associated with progression include apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) mutational signatures, bi-allelic inactivation of tumor suppressor genes together with increased genetic complexity.
Complex events may also be important in progression because they can deregulate more than one gene simultaneously, delivering strong oncogenic signals that may mediate poor clinical outcomes (see Figure 3). We are dissecting out the role played by these structural events in the early phases of disease and their subsequent role at progression.
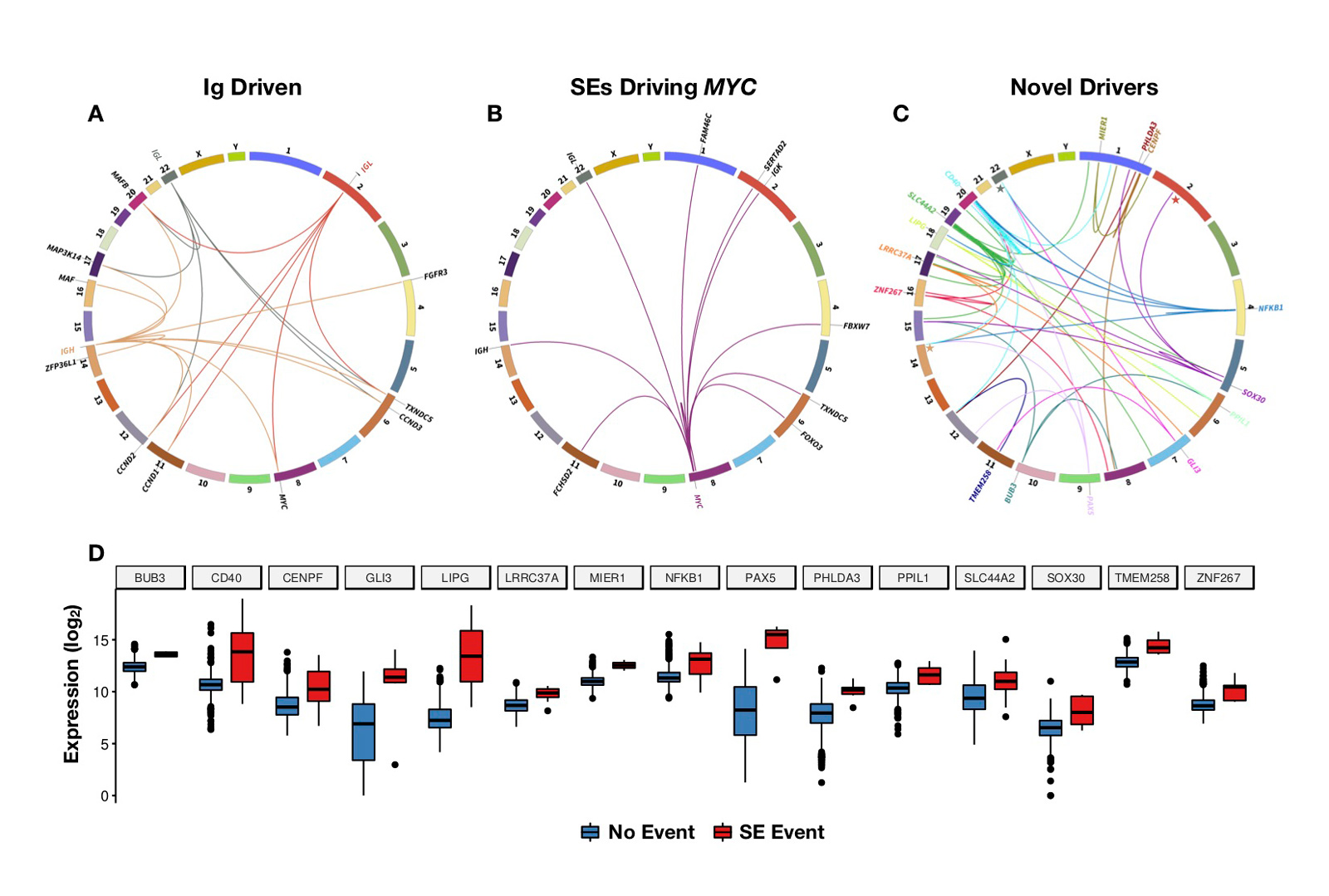
The data we have produced has significantly contributed to the clinical understanding of the biology of progression of myeloma in distinguishing MGUS, smoldering multiple myeloma, and multiple myeloma, in improving prognostic assignment, and finding new factors associated with the risk of developing the disease.
The Molecular Basis and Stage of Origin of Waldenstrom’s Macroglobulinemia
We are applying a similar range of techniques to the analysis of Waldenstrom’s macroglobulinemia as we are using to analyze multiple myeloma. We seek to define the molecular landscape of Waldenstrom’s macroglobulinemia as well as the mutational signatures and evolutionary trajectories leading to disease progression and clinical disease.
The Etiology of Multiple Myeloma and its Excess Risk in African Americans
Genetic predisposition can account for approximatively 10 percent of multiple myeloma cases and with our collaborators we have been able to identify many of the inherited genetic factors contributing to the risk of developing myeloma.
Other factors also contribute including inflammation, aging, obesity, and African American ethnicity, which contributes a three-fold excess risk. Understanding the genetic and environmental contributions to the excess risk will give new insights into the oncogenic pathways leading to multiple myeloma.
To address the risk of developing multiple myeloma by racial origin we are carrying out a whole genome sequencing project of many thousands of cases of myeloma, in patients of different ancestries, to compare the evolutionary trajectories of the early disease phases and whether they differ by racial subgroups.
These comparisons use molecular and immunologic signatures, or “mutagraphs,” that can define both the time of disease origin and the evolutionary trajectories leading to disease progression.
Taking this approach will give insights into the role played by chronic inflammation in both disease initiation and progression.
The concepts of ancestry and ethnicity are different, and we wish to dissect out the contribution of each to the risk of developing multiple myeloma. We will address this question by using a genetic admixture tool to accurately assign racial origin.
To gain further insights, we will integrate environmental information collected as part of a clinical trial of smoldering myeloma. This study is focused on patients of African American origin and will integrate clinical, environmental, and socio-economic information. The study will use sequential analysis, epidemiologic data, and immune and microbiome analyses.
The Molecular Basis of Clinically Aggressive High-Risk Behavior
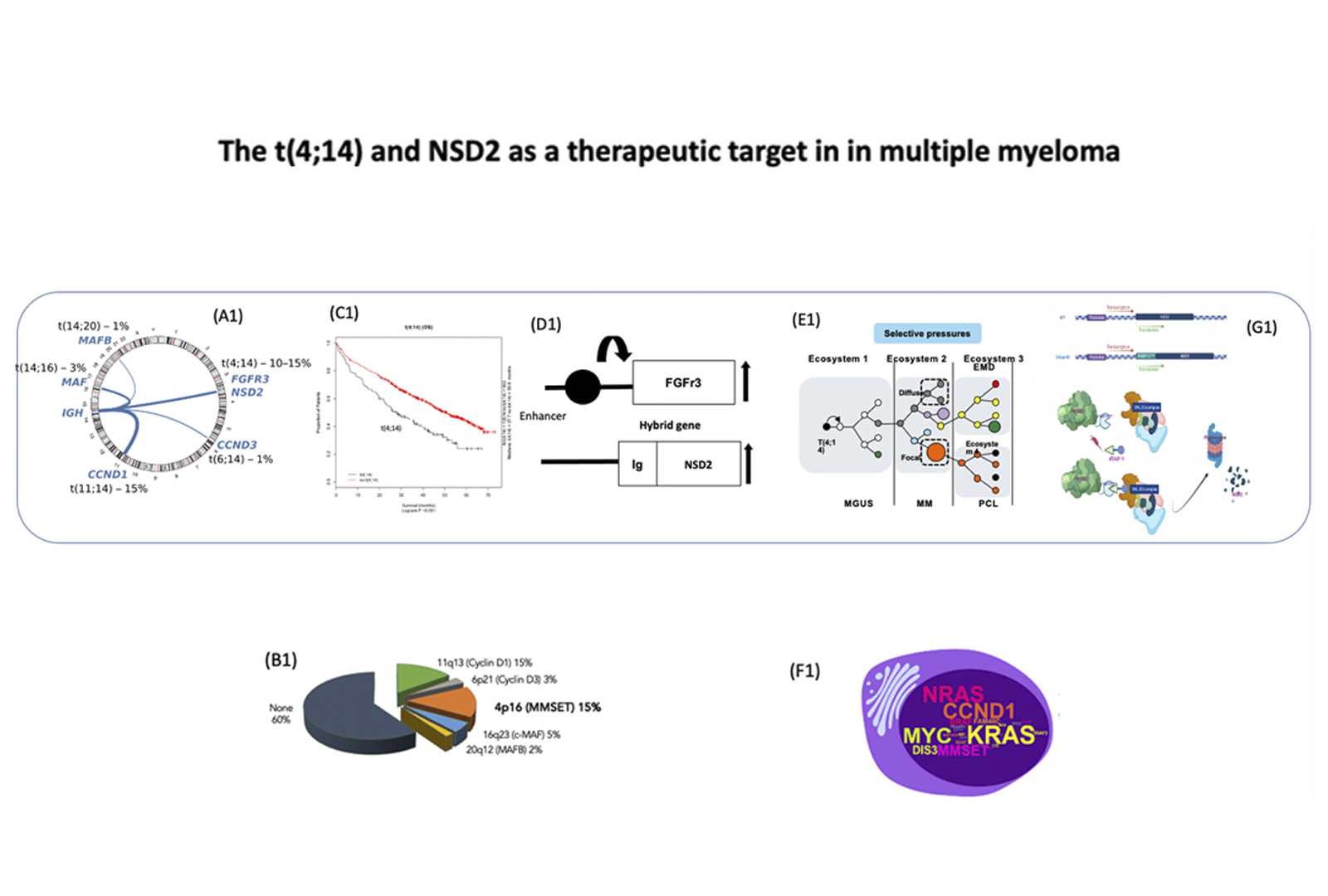
We have a particular interest in drivers of aggressive clinical behavior caused by genetic events such as the t(4;14) and amplification of the long arm of chromosome 1. We are addressing how the deregulation of the expression and function of key epigenetic modifiers and transcription factors affects disease progression and contributes to high-risk disease.
NSD2
The t(4;14) has been characterized at a molecular level and directly leads to the upregulation of the histone methyl-transferase NSD2. We have set up a number of model systems, including CRISPR knockout and the dTAG systems, to address the role played by NSD2 on the chromatin landscape using multiomic technologies. This information that will allow us to understand its role in t(4;14) myeloma and potentially expand our capacity to target it (see Figure 4).
PHF19
We have shown that one of the prognostic features for aggressive clinical disease is the overexpression of PHF19, an epigenetically-active Tudor Domain containing protein. PHF19 is part of the PRC2 complex which may have a key role in aggressive disease and we are investigating this role further in model systems.
Gain of Chromosome 1
One of the key players in aggressive disease is the gain and amplification of chromosome 1. In order to define the drivers on chromosome 1 we have studied chromosome 1 at a structural level and will take a genomics approach to study important regulons located along the long arm and their impact on chromatin.
The Tumor Microenvironment and Aggressive Myeloma
We are addressing the interaction between genomic factors, the microenvironment, and response to treatment. Using simultaneous single-cell epitope and transcriptome measurement of both the tumor cells and the microenvironment, we have identified a “high-risk microenvironment”. This environment is characterized by an excess of classical monocytes that differ between responders and non-responders. Our future analyses will focus on cellular and chromatin states to better understand the effect of the malignant plasma cells on the monocyte compartment in vitro.
Targeted Therapeutics for Multiple Myeloma
We are collaborating with other investigators at NYU Langone to investigate novel targeted therapeutic strategies for multiple myeloma. In particular, we are looking at the role of targeted nanobody conjugates and targeting the endoplasmic reticulum as a means of therapeutically addressing RAS mutated myeloma.