
Stem Cell Dynamics: In Vivo & In Silico
Unlike somatic cell lineages in C. elegans, cell divisions within the germ line are more like mammalian tissues in that they do not occur in a reproducible pattern. Thus, the spatial and temporal dynamics of germ cell division within the proliferation-competent zone are not well defined (Hubbard, 2007). No current markers exist to exclusively label putative stem cells, the cell cycle in the mitotic zone is relatively slow, and it is not yet feasible to follow multiple divisions over a suitable time frame in live animals. Hence, the exact mechanism for renewal in this otherwise highly tractable system is unknown, impeding optimal use of the system.
Over the years investigators in the E. Jane Albert Hubbard Lab have sought to circumvent these limitations. We took a wet/dry quantitative approach to investigate the dynamics of cell division in the adult proliferation zone (Maciejowski et al., 2006). We collaborated with computer scientists in the group of David Harel at the Weizmann Institute (Fisher et al., 2005; Kam et al., 2008; Sadot et al. 2008). In a recent project with the group of Hillel Kugler, now at Microsoft Research in Cambridge, England, we extended the work to model germline dynamics. We built several versions of a model that recapitulated key aspects of germline progenitor pool dynamics and experimentally validated several surprising observations made from model-driven simulations (Setty et al., 2012; Atwell et al., 2015).
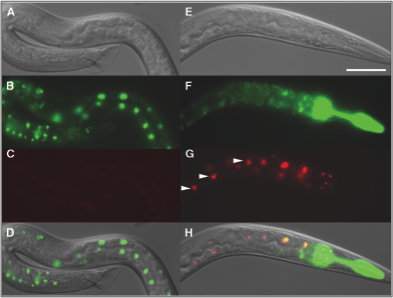
In parallel, we are developing experimental technologies. We adapted the FLP-out system for use in C. elegans and demonstrated that single somatic cells can be marked and their progeny traced using this technique (Voutev and Hubbard, 2008). For germline studies, we are adapting this system and other cell-marking and lineage-tracing systems to mark stem cells and their progeny. This work is greatly facilitated by new technologies in the field that enable germline expression of transgenes as well as precise genome editing. Together, these tools (modeling, lineage tracing, and live imaging) will enable us to exploit this system fully and provide an unprecedented link between the effects of genetic and environmental perturbations on specific subsets of cells and overall cell dynamics within the stem cell population.
Relevant Publications
Atwell K … Hubbard EJ. How computational models contribute to our understanding of the germ line. Mol Reprod Dev. 2016. DOI.
Atwell K … Osborne JM. Mechano-logical model of C. elegans germ line suggests feedback on the cell cycle. Development. 2015. DOI.
Setty Y … Kugler H. A model of stem cell population dynamics: in silico analysis and in vivo validation. Development. 2012. DOI.
Voutev R and Hubbard EJ. A "FLP-Out" system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008. DOI.
Kam N … Hubbard EJ. A scenario-based approach to modeling development: a prototype model of C. elegans vulval fate specification. Dev Biol. 2008. DOI.
Sadot A … Harel D. Toward verified biological models. IEEE/ACM Trans Comput Biol Bioinform. 2008. DOI.
Hubbard EJ. Caenorhabditis elegans germ line: a model for stem cell biology. Dev Dyn. 2007. DOI.
Maciejowski J … Hubbard EJ. Quantitative analysis of germline mitosis in adult C. elegans. Dev Biol. 2006. DOI.
Fisher J … Harel D. Computational insights into Caenorhabditis elegans vulval development. Proc Natl Acad Sci. 2005. DOI.