
Insulin Receptor
Using X-ray crystallography as our primary experimental technique, researchers in the Hubbard Lab are attempting to understand the molecular basis for insulin receptor activation and for recruitment of downstream signaling proteins to the activated (phosphorylated) insulin receptor (see figure 1).
Several cytoplasmic adapter proteins bind to the activated insulin receptor, including insulin receptor substrate (IRS) proteins and APS, which are positive factors in insulin signaling pathways culminating in glucose uptake. The insulin receptor is downregulated by the adapter proteins Grb14 and Grb10 as well as the tyrosine phosphatase PTP1B.
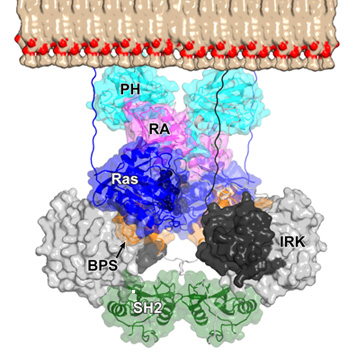
Read more in our paper “Structural and functional studies of the Ras-associating and pleckstrin-homology domains of Grb10 and Grb14” published in Nature Structural & Molecular Biology.
We are determining crystal structures of complexes between these proteins and the insulin receptor kinase domain to elucidate the modes of interaction and the determinants of specificity (see figure 2).
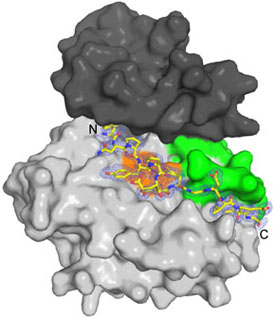
Read more in our paper “Structural and biochemical characterization of the KRLB region in insulin receptor substrate-2” published in Nature Structural & Molecular Biology.