
B-cell Superantigens in Health & Disease
For many years the B-cell Immunobiology Lab pioneered investigations of the molecular pathways responsible for the properties of B-cell superantigens. We should first explain the molecular basis of B cell recognition of conventional antigens, as the general structural and genetic principles that underlie conventional antigen recognition by an antibody are well known. In general, the binding of an antigen involves contacts from both the heavy-chain variable region (VH) and the light-chain variable region (VL). In each V region, there are three non-contiguous linear intervals of greatest variability, which have been termed hypervariable regions or complementarity-determining regions (CDRs). Separating these CDRs are more highly conserved intervals termed framework regions. In the β-barrel structure of an antibody, the framework regions fold into relatively rigid β strands that maintain the overall antibody structure, whereas the CDRs form β bends (or loops) that are juxtaposed to form a composite surface at one end of the Fab to provide the contacts for antigen. The recognition of an antigen requires a combinatorial interaction of residues from two or more of these loops, with contributions that always include the somatically generated CDR3 subdomains.
What is a B-cell superantigen?
Superantigens are microbial toxins that interact with immune receptors of B lymphocytes (B cell receptors, BCR) outside the usual sites involved in the recognition of antigens e.g. complementary determining regions (CDRs), to elicit a response within the target cell. The interaction of a B cell superantigen with a BCR is a non-immune interaction that is believed to have emerged during the co-evolution of the microbe and the mammalian host. A toxin with superantigen properties developed by a microbe to confound and subvert host immune defenses.
Whereas conventional antigens generally interact with less than 0.1% of the naive lymphocyte repertoire, based on unconventional binding interaction outside of the antigen binding pocket, a superantigen can have BCR-mediated interactions with a large proportion of the lymphocyte pool (often more than 5%).
Understanding how a bacterial protein could induce BCR-mediated B cell activation on such a supraclonal scale was, therefore, quite an enigma. Earlier attempts to elucidate the structural basis for Ig binding to the Fab-mediated “alternative binding site” of SpA using single point mutagenesis of BCR were less than satisfying (Sasso et al. 1989; Sasso et al. 1991). The mystery was solved by the first 3-D structure for a B-cell superantigen, using a single domain of SpA, in complex with the VH3-encoded Fab from a human rheumatoid factor. This revealed that binding was specific for virtually all products of the genes of the VH3 gene family (the human representative of the mammalian VHIII clan that is integral to the immune systems of almost all mammalian species. This contact site is a composite conformational solvent-exposed surface on the outside of the Fab, formed by contributions of VH framework 1 and framework 3 subdomains (Graille et al. 2000), which reiterates general themes first documented for microbial toxins with properties of T cell superantigens (reviewed by (Deacy et al. 2021)).
Using the prototypic superantigen, Staphylococcal protein A (SpA), as a model system to understand how different B-cell antigen receptor encounters lead to lymphocyte activation, proliferation, or apoptotic deletion, we have studied a class of natural microbial toxins that we have demonstrated can act as superantigens affecting large sets of B-cells, defined by their antibody variable region gene usage.
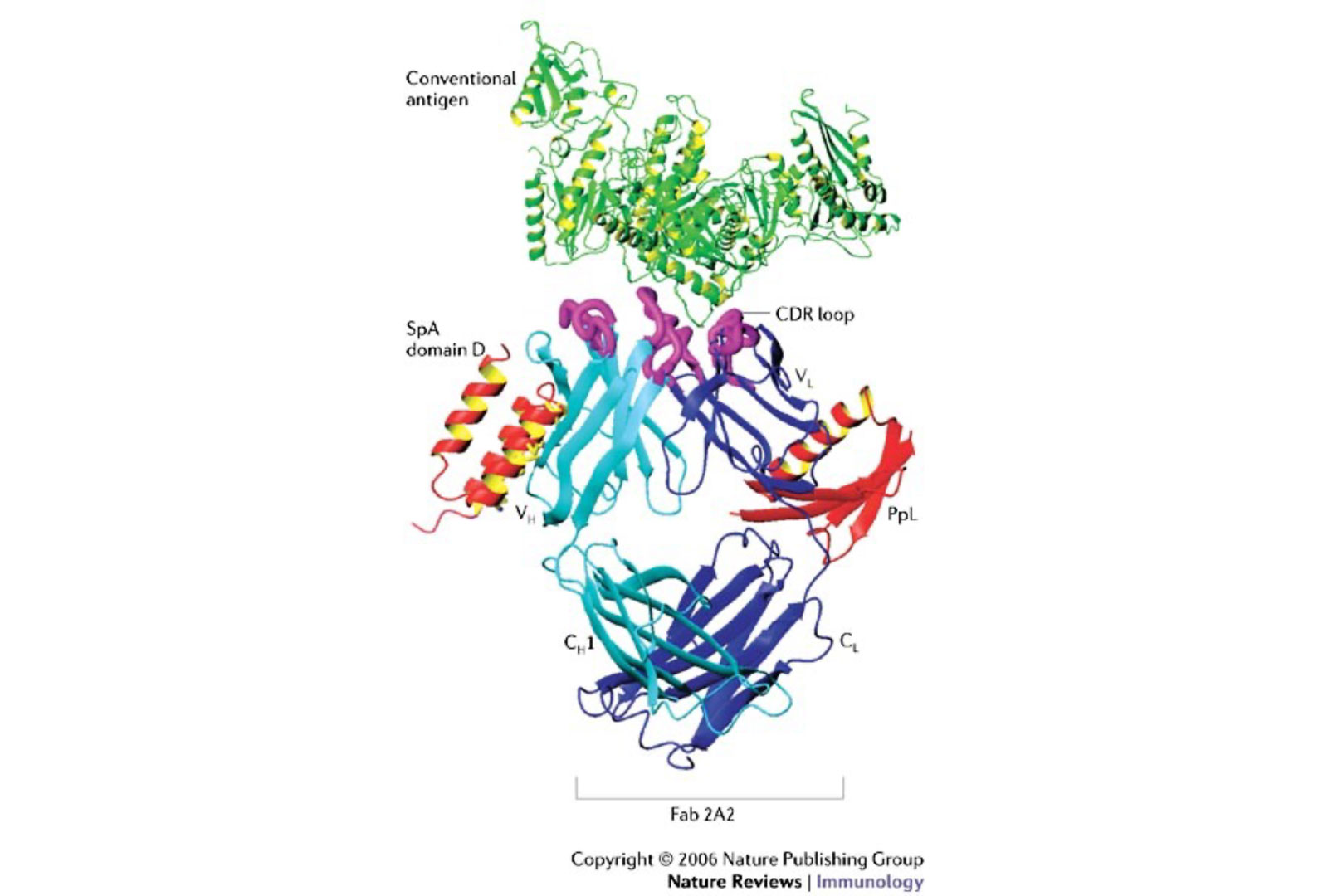
Over the years, we have studied a class of natural microbial toxins that we have demonstrated can act as superantigens affecting large sets of B-cells, defined by their antibody variable region gene usage. We have used the prototypic superantigen, Staphylococcal protein A (SpA), as a model system to understand how different B-cell antigen receptor encounters lead to lymphocyte activation, proliferation, or apoptotic deletion.
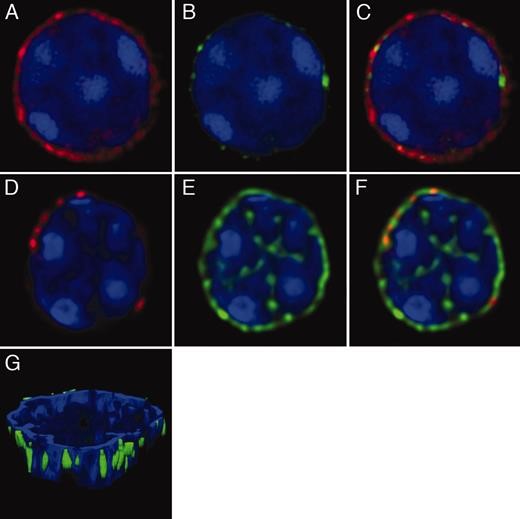
Our lab has shown how in vivo encounters with SpA (see Figure XX), can result in targeted activation-induced cell death. We also identified the exact intrinsic pathway of apoptosis and demonstrated the requirement for a single pro-death Bcl-2 family member, Bim. Parallel studies are in progress to assess similarities and differences for CD20-mediated B-cell deletion occurring after treatments with the recombinant antibody to the CD20 antigen specifically expressed on the surface of B lymphocytes, Rituxan. These studies are relevant to understanding and developing better therapeutic agents for the selective targeting and deletion of malignant B-cell populations in models of lymphoma and leukemia.
We wondered whether exposure to a bacterial superantigen can induce the death of a lymphocyte by intracellular pathways akin to those also detected in B cells dying from treatment with Rituximab, which is the archetype anti-CD20 antibody that acts predominantly by antibody-dependent cytotoxicity (ADCC). For these studies, we used the T15i VH transgenic mouse of Rajewsky (Taki et al. 1993), which expresses a monoclonal VH3-related H chain in combination with diverse VL regions on a C57BL/6 background. After in vivo exposure to SpA, which contains five Fab binding domains in tandem that can cross-link B-cell membrane-associated Fab of the VHIII clan, these B cells are susceptible to Activation Induced Cell Death (AICD).
In the B-cell Immunobiology Lab, we are investigating how microbial B-cell superantigens affect in vivo B-cell repertoires (see Silverman and Goodyear, Nature Reviews in Immunology, 2006). As shown below (see figure 1), we reported the first B-cell repertoire surveys using Next-Generation DNA sequencing (NGS) that showed the capacity of exposure to prototypic microbial B-cell superantigen, Protein L (PpL) made by Peptostreptococcus magnus, which was shown to induce global shifts in the in vivo representation of the B-cell repertoire (Grönwall et al., 2012, Journal of Immunology) (see figure 1). For these analyses, we worked with Prof. Sergei K. Pond (Temple University, formerly at UC San Diego) and colleagues to develop a high throughput computational pipeline for the analysis and visualization of the composition of human immune repertoires (see Frost et al., 2015, Philos Trans R Soc Lond B Biol Sci).

As described above, while in earlier investigations we had characterized the cellular and molecular properties of archetypic microbial B cell superantigens, such as PpL and SpA, it was unexpected that the bacterial commensal that identified as an overrepresented pathbiont, Ruminococcus (Blautia) gnavus, in Lupus nephritis (Azzouz et al., 2019, Ann Rheum Dis), was recently found to release a distinct B-cell superantigen into the gut of colonized humans. Sequence analysis indicates that the properties of this microbial factor, which are akin to those of SpA, arose by convergent evolution. In our current studies, we are in part investigating how this factor may contribute to the pathogenesis of clinical lupus nephritis.