
Microbiome & Systemic Lupus Erythematosus
Systemic lupus erythematosus (SLE), the archetypal systemic autoimmune disease, is believed to arise due to the influence, in combination, of genetic and environmental factors. Several recent studies have shown that systemic inflammatory and autoimmune diseases may arise in mice and humans from imbalances in the commensal microorganism community, which exists in a mutualistic relationship within the gastrointestinal tract. While genetic defects and other environmental factors may predispose some individuals to immune-mediated diseases, we hypothesized that pathogenic symbiont bacteria (termed pathobionts) among the commensal intestinal microbiota may be the direct trigger(s) for the SLE pathogenesis.
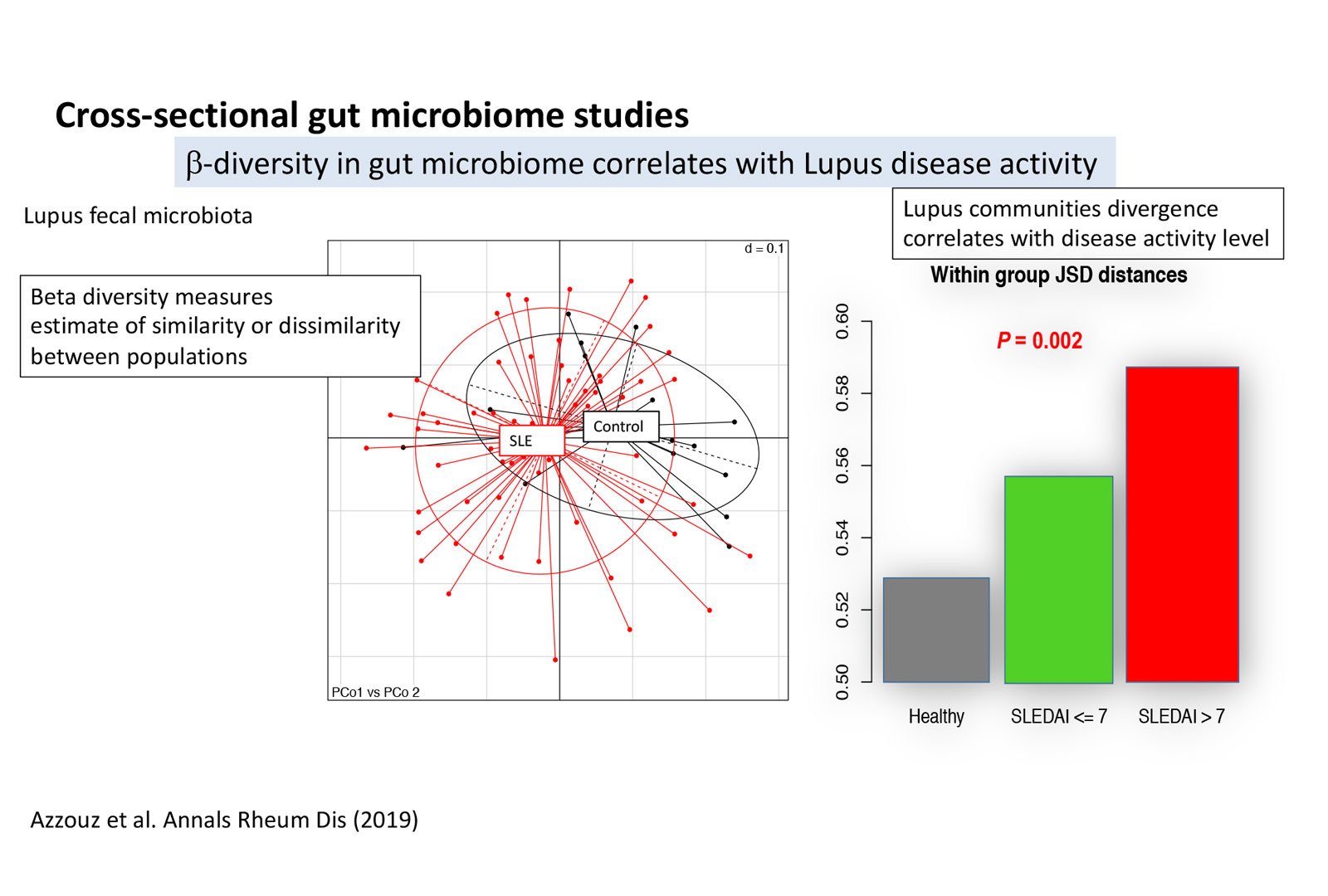
In collaboration with Drs. Buyon and Izmirly of the Division of Rheumatology, the B-cell Immunobiology Lab has recruited a new cohort of SLE patients from NYU urban clinics. We performed surveys of the intestinal microbiota of our SLE cohort, with Next-Generation Sequencing of the 16S bacterial rRNA gene in gut communities. Analysis of these data revealed that SLE patients displayed decreased richness or diversity in their microbiome communities, with reductions in taxonomic complexity most pronounced in those with high SLE disease activity index (SLEDAI) compared to the healthy control (HCTL) subjects (p<0.002). SLE subjects also demonstrated greater intra-group diversity (p<0.005) than did the healthy female controls.
Strikingly, SLE patients with high SLEDAI scores had an overall five-fold greater representation of Ruminococcus gnavus (RG) of the Lachnospiraceae family (p=0.001, Figure 1A), this bacterial species was most highly represented in patients with active renal disease, which, in turn, raised our attention to RG as a possible causative pathobiont in lupus pathogenesis. (Figures 1–3, see Azzouz et al., 2019, Annals of the Rheumatic Diseases.)
Image (below): Cross-Sectional Gut Microbiome Studies
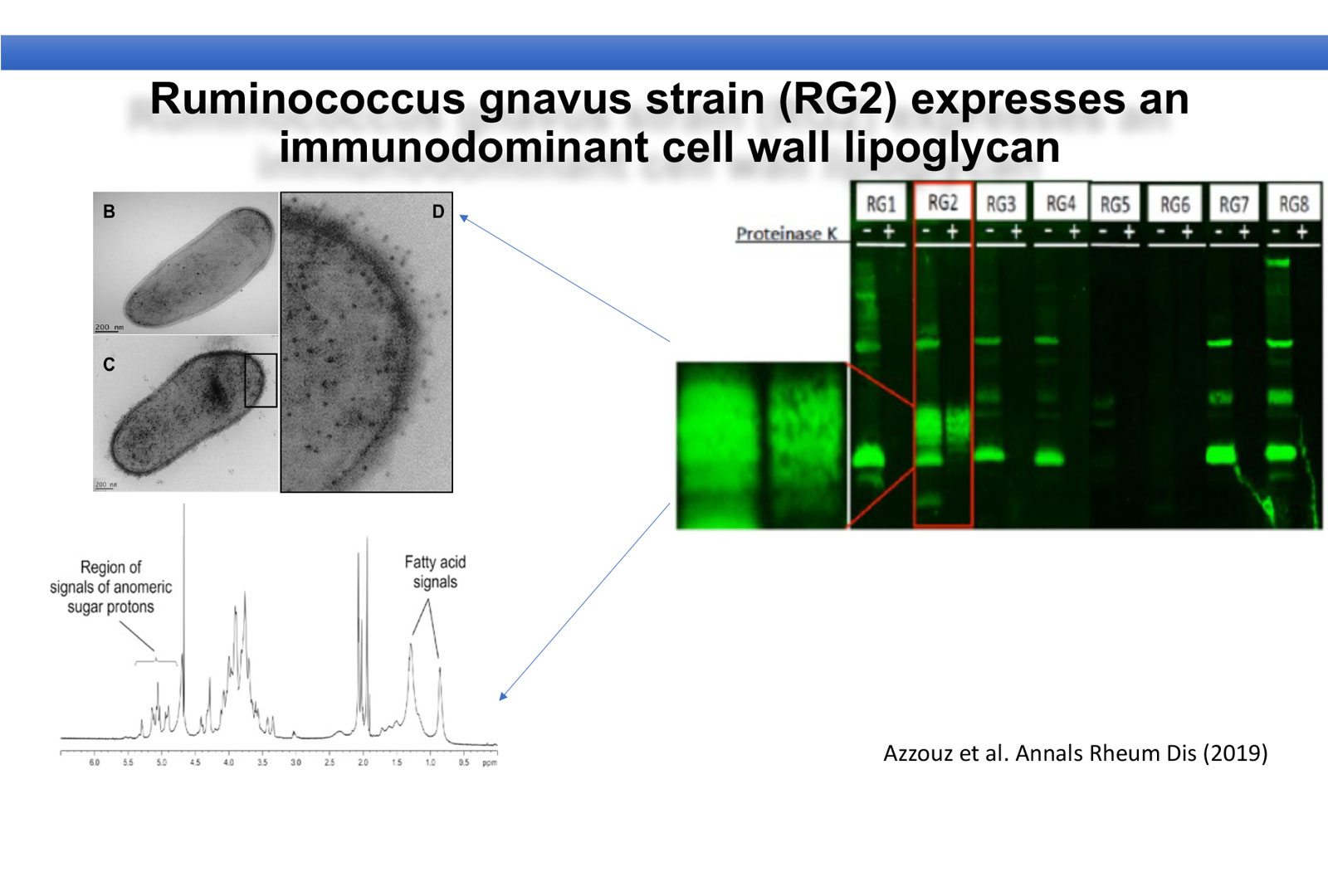
To understand the link between RG expansion and SLE pathogenesis, we first questioned whether the immune systems of SLE patients recognize antigens produced by different strains of RG. Toward this goal, we evaluated the reactivity of IgG antibodies in the sera of SLE patients against eight different strains of RG obtained from different donors. At right, in a representative immunoblot using the serum of a SLE patient with active Lupus nephritis and high intestinal representation of RG, we found a high level of IgG antibodies against a distinct antigen in only one strain (RG2), which was not commonly found in the sera of healthy controls (not shown). This IgG reactivity correlated significantly with high lupus disease activity based on the SLEDAI score, with gut abundance of RG, and with serum levels of anti-dsDNA IgG (a biomarker of lupus renal disease)(not shown). This RG2 antigen was not degraded by digestion with a protease, which suggested this was a non-protein bacterial glycan.
This immune recognition of the distinct RG2 strain-associated antigen by SLE serum IgG (Upper left) is directed against a cell wall/lipoglycan antigen in the RG2 strain. We used an electron microscopy approach to detect and visualize the IgG-specific binding to this glycan structure that showed binding on the bacterial cell surface. Structural chemical analysis of H-NMR and Mass Spectroscopy, showed signals of an oligosaccharide and fatty acid indicative of a cell wall-associated lipoglycan structure (and not a teichoic acid) that is an immunodominant immunogenic antigen on the RG2 strain (in collaboration with N. Gisch and co-workers at the Borstel Institute Germany). By performing in vitro stimulation assays of a reporter cell line, we discovered that this lipoglycan is a TLR2 ligand and has a potent immune modulator role that is inhibited in a dose-dependent fashion by monoclonal antibodies that are specific for human TLR2 that acts as a receptor on the cell line (not shown).
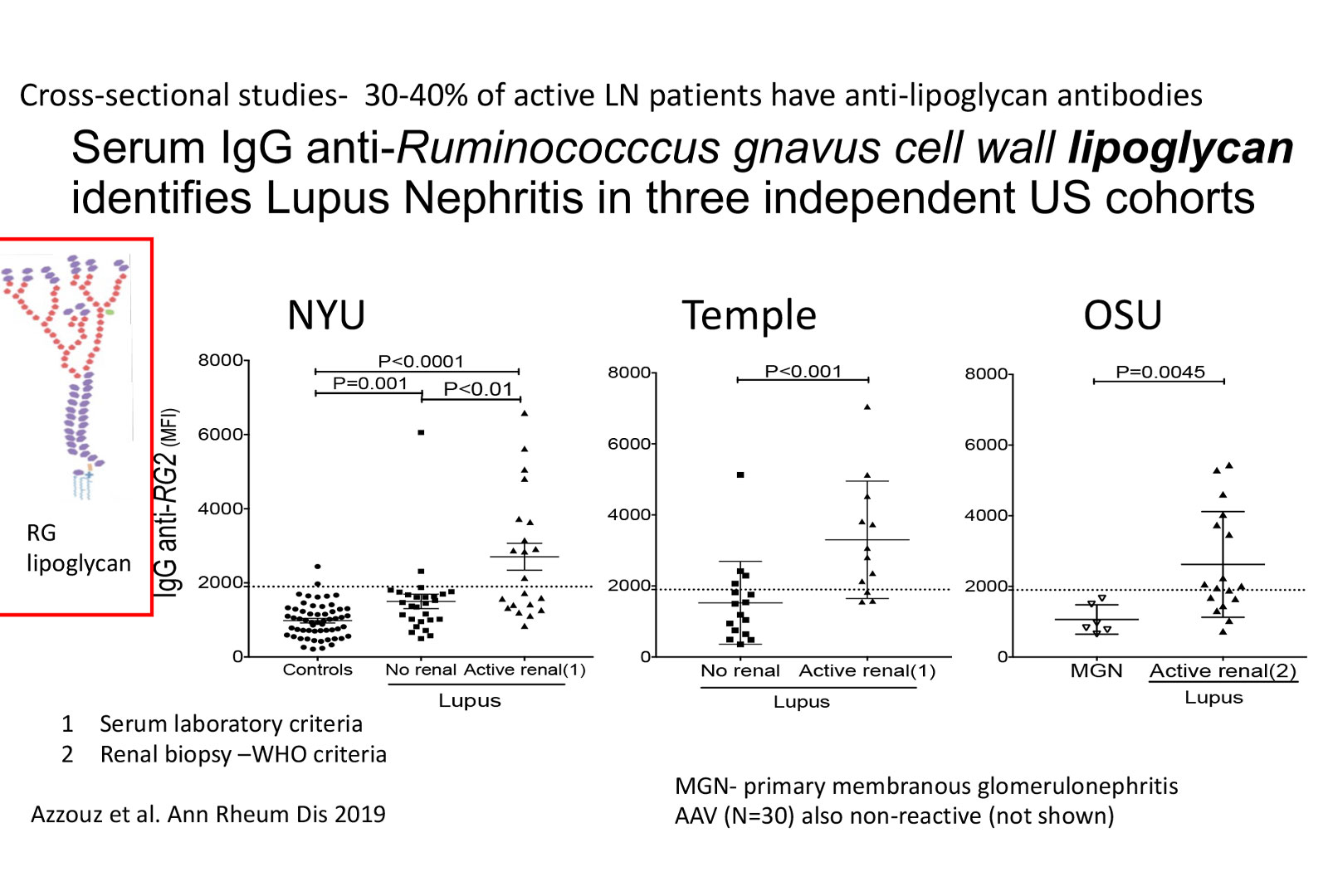
These findings were validated in three independent Lupus patient cohorts at other collaborating academic centers
We are currently developing approaches to test whether we can target SLE-associated imbalances in the gut microbiome. More specifically, we postulate clinical benefits will result if we target the RG pathobiont bacteria, and prevent host systemic immune responses to it. We are also currently evaluating whether the serum antibody response to the RG bacteria can be used as a prognostic marker, and as a predictor in SLE patients of developing renal involvement.
Supported by the Lupus Research Institute, the Judith and Stewart Colton Center for Autoimmunity, and the National Institute of Arthritis and Musculoskeletal and Skin Diseases.