Multi-Omics Study Design & Data Integration Resource Resources
NYU Langone’s Multi-Omics Study Design and Data Integration Resource (MOSDIR) provides researchers with a comprehensive suite of resources for multi-omics data analysis. To foster knowledge sharing, MOSDIR also hosts a monthly journal club to explore cutting-edge advancements in the field. Resources are continuously updated and expanded to maintain their relevance and utility.
Analytic Resources
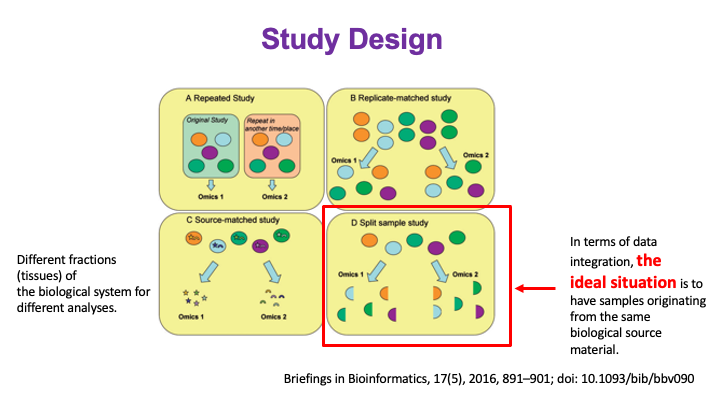
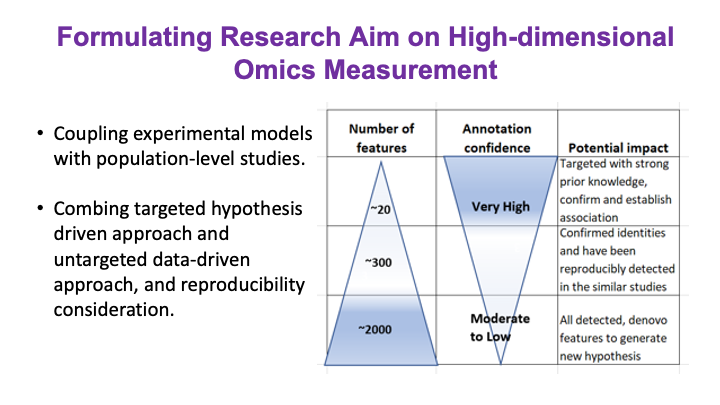
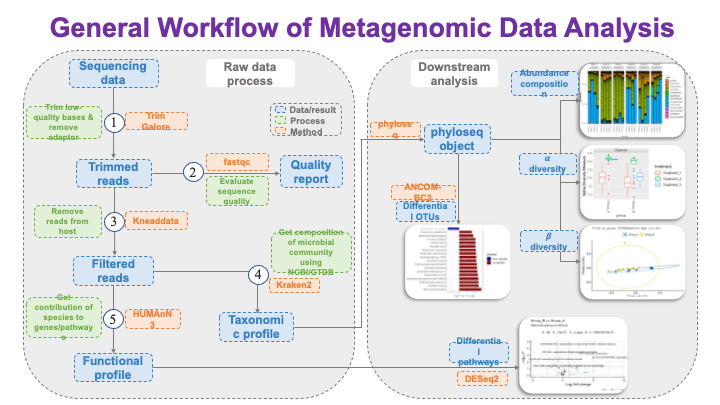
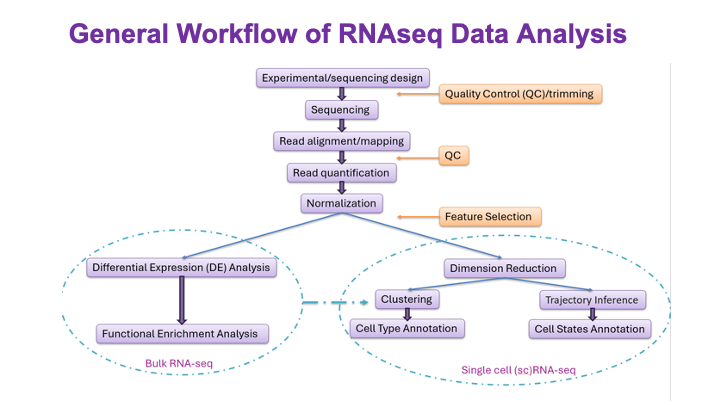
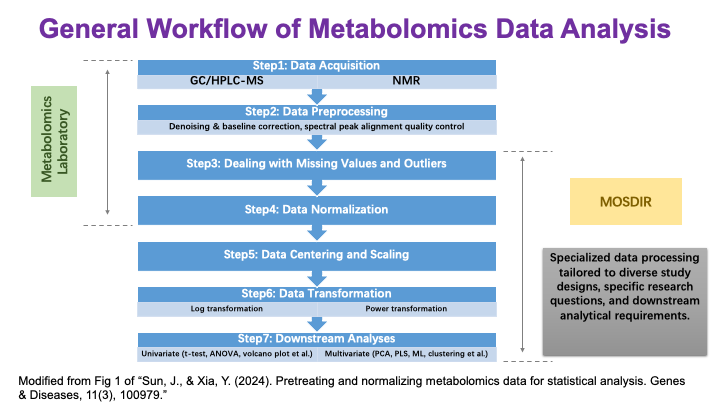
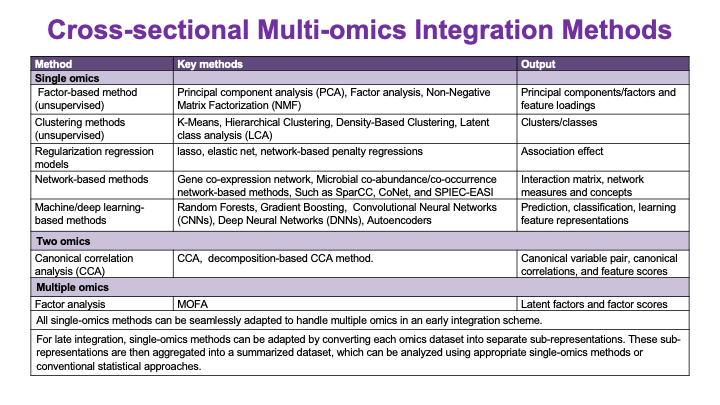
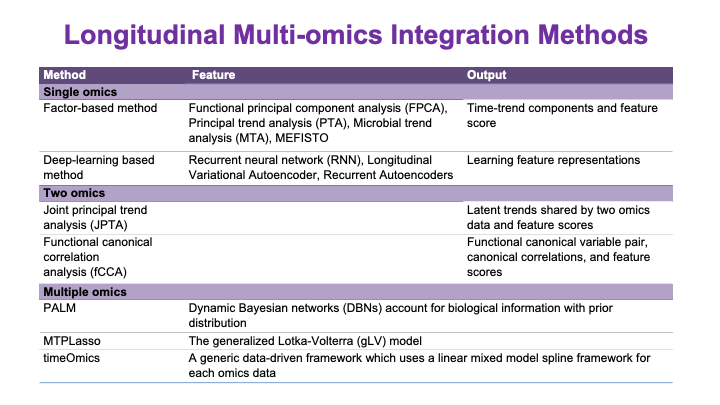
These slides provide an overview of MOSDIR's analytical resources, highlighting guidance on study design, protocols for processing and analyzing single-omics data (e.g., metagenomics RNA-seq, and metabolomics), and methodologies for integrating multi-omics data in both cross-sectional and longitudinal studies.
MOSDIR Image Gallery
Journal Clubs
The Multi-Omics Data Integration Journal Club convenes monthly to review a significant multi-omics paper, promoting our knowledge and keeping us up to date with the latest advances in the field. Typically, our Journal Club meetings are held on the fourth Friday at 11am of each month during academic semesters. We offer a hybrid format, enabling participation either in person or via Webex. To express your interest in joining, please reach out to us at Huilin.Li@NYULangone.org, and we will promptly provide further details.
In our recent sessions, we've studied the following papers:
- Simner PJ and Salzberg SL (2022). The human “contaminome” and understanding infectious disease. New England Journal of Medicine. 387(10):943-6.
- Gihawi A, et al (2023). Major data analysis errors invalidate cancer microbiome findings. MBio. 14(5):e01607-23.
- Velten B and Stegle O (2023). Principles and challenges of modeling temporal and spatial omics data. Nature Methods. p.1462-74.
- Vasaikar SV, et al (2023). A comprehensive platform for analyzing longitudinal multi-omics data. Nature Communications. 14(1):1684.
- Wang FA, et al (2024). TMO-Net: an explainable pretrained multi-omics model for multi-task learning in oncology. Genome Biology. 25(1):1-24.
- Tang X, et al (2023). Explainable multi-task learning for multi-modality biological data analysis. Nature communications, 14(1), p.2546.
- Diray-Arce J, et al (2023). Multi-omic longitudinal study reveals immune correlates of clinical course among hospitalized COVID-19 patients. Cell Reports Medicine, 4(6).
- Arıkan M and Muth T (2023). Integrated multi-omics analyses of microbial communities: a review of the current state and future directions. Molecular Omics.
- Lin H and Peddada SD (2024). Multigroup analysis of compositions of microbiomes with covariate adjustments and repeated measures. Nature Methods, 21(1), pp.83-91.
- Singh A, et al (2019). DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 35(17):3055-62.